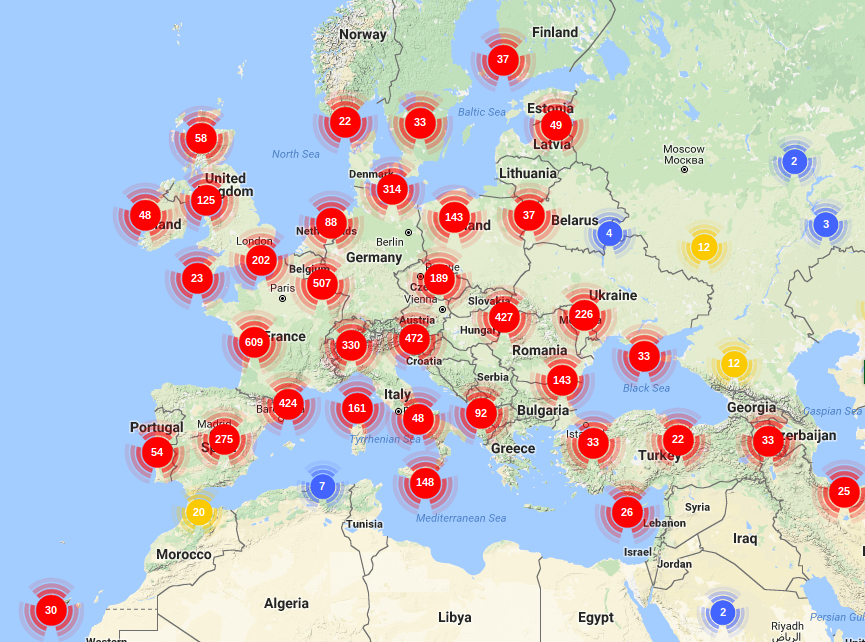
PHYLOGENY
Phylogeny based on morphological traits
The first cladistics analysis of the genus Quercus relying on morphological characters is by Nixon (1993a) and is based on the evolutionary variation of the pistillate flower and fruit subtending the cupule (Kaul and Abbe 1984). Phylogenetic relationships based on morphology could be proposed once polarization of flower and fruit traits were inferred from the existing fossil remains (Nixon 1989, 1993a). Morphological synapomorphies of the genus Quercus include a single pistillate flower, valveless cupules, decurrent styles, expanded stigmatic surfaces, unisexual inflorescences, lax staminate inflorescences and scabrate pollen exine structure (Manos et al., 2001). Two subgenera were recognized by Nixon (1993a): Cyclobalanopsis (cycle-cup oaks, only present in Asia) and Quercus (oaks, widespread over the 3 continents).
Subgenus Quercus can be distinguished from sub genus Cyclobalanopsis by the presence of expanded stigmatic surfaces on the pistillate flowers and small inconspicuous bracts which subtend single-staminate flowers.
Within subgenus Quercus, Nixon recognized three clades:
- Section Lobatae (= Erythrobalanus, sensu Camus, red and black oaks, present only in North and Central America) showing most of the plesiomorphic morphological traits within Subgenus Quercus. This section is defined by a derived pistillate floral morphology (a calix “flange”) that is absent in the rest of Quercus (Nixon, 1993b).
- Section Protobalanus (= Protobalanus, sensu Camus, intermediate or golden cup oaks, present only in North America), with abortive ovules apical to lateral.
- Section Quercus (= Lepidobalanus, sensu Camus, white oaks, present in the three continents), with abortive ovules always basal. Because of the synapomorphy of the basal position of the abortive ovules on the surface of the development seed, this section includes also two groups that were identified in previous classifications and comprise many plesiomorphic characters: the Cerris and Ilex group.
More recently pollen features as patterns of tectum ornementation assessed by electronic microscopy allowed to resolve morphological phylogenetic relationships within section Quercus (Denk and Grimm, 2009). Tectum ornamentation can be used to polarize character states within Quercus by comparison with other genera in the Fagaceae. Species of the Ilex group exhibit distinct simple type tectum ornamentation also found in Fagus and in extinct lineages of Trigonobalanus, Colombobalanus and Formamendron.
Phylogeny based on molecular data
Phylogenetic reconstructions were further enriched with molecular data coming from various sources:
- Chloroplast DNA restriction analysis: Manos et al. (1999)
- Chloroplast DNA sequence data (rbcl, atpBE intergenic spacer;3’ trnK intron): Manos and Stanford (2001)
- Chloroplast DNA sequence data (intergenic spacer trnD-trnT): Xu (2004)
- Ribosomal sequences (ITS region): Samuel et al. (1998), Manos et al. (1999), Manos et al. (2001)
- Single copy nuclear gene CRABS CLAW (CRC), that regulates carpel development in angiosperms: Oh and Manos (2008)
Analysis of the combined cpDNA and ITS data were more resolute and ended into four monophyletic groups (sensu Camus): (Cerris-(Erythrobalanus-Protobalanus+Lepidobalanus); the same resolution was obtained within cpDNA sequence diversity within the interspacer trnD-trnT (Xu, 2004), although this analysis additionally separated sub section Brachylepides at a higher hierarchical level: [Sub section Brachylepides–(Cerris (Erythrobalanus + Protobalanus + Lepidobalanus)]
One of the most complete analyses based on the nuclear sequences (CRC and ITS) resolved two clades with strong support:
- The Eurasian Cerris clade, that is actually closely related to the subgenus Cyclobalanopsis.
- The mostly new world clade composed of Erythrobalanus-Protobalanus-Lepidobalanus.
They suggest that among the 6 botanical sections identified by Camus (Erythrobalanus, Lepidobalanus, Protobalanus, Cerris, Mesobalanus, and Macrobalanus), only the four first are distinguished as monophyletic groups in the phylogenetic reconstruction. Species assigned to Mesobalanus, and Macrobalanus by Camus are members of the Lepidobalanus group (Xu, 2004). Two new sections should however be added based on the phylogenetic analysis: the Ilex group, which is part of the Lepidobalanus section in Camus’ classification, and the Brachylepides subsection (sensu Camus). Species of this subsection are located on high altitudes on the eastern side of the Himalayas.
Concerning Nixon’s classification (1993a), the three groups (Quercus, Cerris and Ilex) that were assembled within section Quercus appear as three different clades in the phylogenetic reconstructions and should be considered as three separate sections.
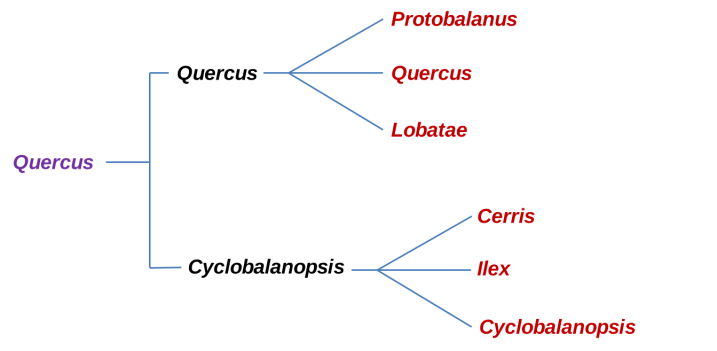
Phylogeny of the major clades of Oaks before RADseq analysis. In Violet is the Quercus genus, Black is the Sub-genus and in Red the Sections.
In view of the problems of resolution encountered with plastid sequence data, oak phylogeneticists concentrated on nuclear-encoded sequence regions, establishing the ITS phylogeny with single-copy nuclear gene regions, multicopy nuclear rDNA and RADseq (Manos et al., 1999; Oh and Manos, 2008; Denk and Grimm, 2010; Hubert et al., 2014). All these analyses support it the recognition of two reciprocally monophyletic groups that can be formalized as two subgenera with eight phylogenetic lineages, which in turn are sections that match the morphological groups determined first by Trelease (1924).
However, the current results also indicate that subsections/series recognized in the monographs of Trelease, Camus, and Menitsky do not always correspond to groups identified using molecular sequence data. These mismatches between traditional classification and DNA-based groups are found in many infrageneric groups and pose a major challenge when searching for morphological criteria to subdivide sections within oaks (Hubert et al., 2014).
Recent molecular phylogenetic studies (Denk & Grimm, 2010; Hipp et al. 2014, 2015, 2017, 2018; Hubert et al., 2014; Manos et al., 2001; McVay et al. 2017; Oh & Manos, 2008) consistently suggest two major clades within oaks, one comprising three Old World groups (sections Cyclobalanopsis, Ilex, and Cerris), the other comprising three New World groups (sections Protobalanus, Virentes, and Lobatae,) and two northern hemispheric groups (sections Ponticae and Quercus).
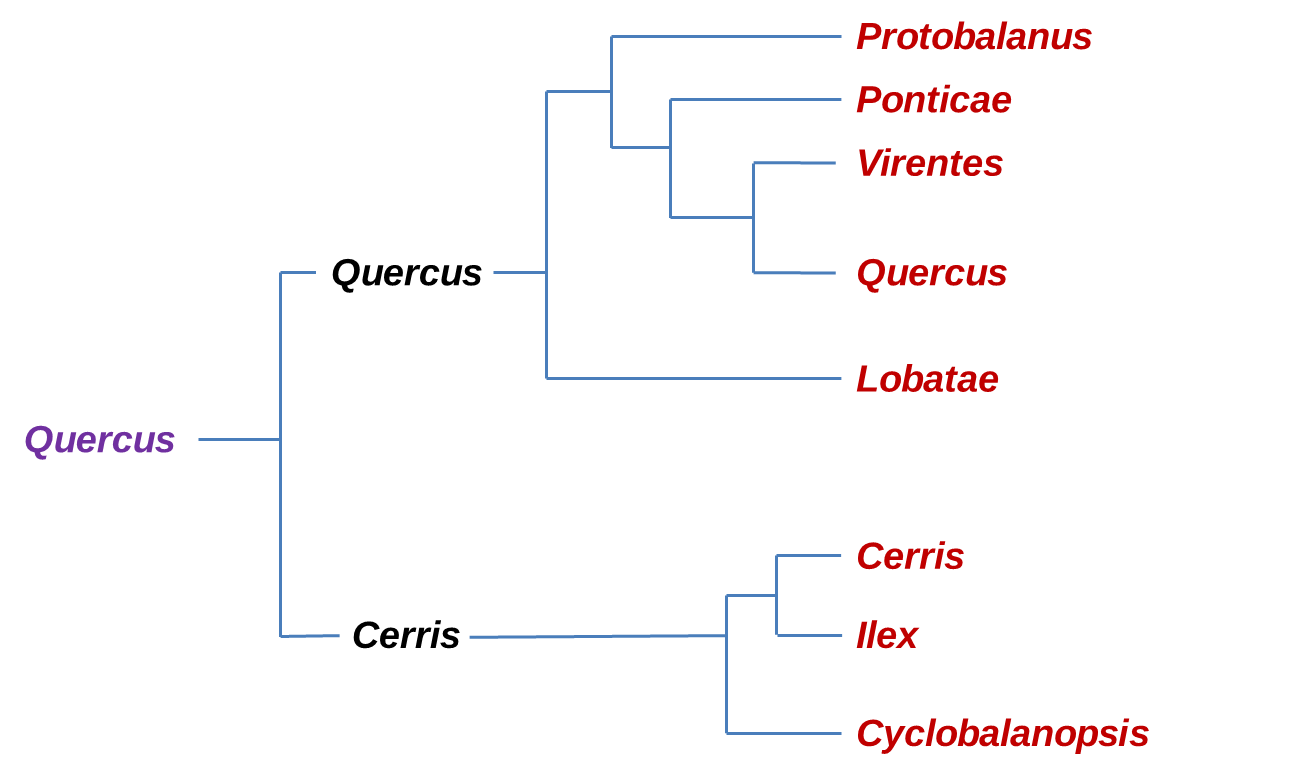
Phylogeny of the major clades of Oaks according to the new classification by Denk et al (2017) confirm by RADseq (McVay et al., 2017). In Violet is the Quercus genus, Black is the Sub-genus and in Red the Sections.
The section Ponticae is represented by only two species, Quercus pontica (in Western Eurasia) and Quercus sadleriana (in the West of North America) which are widely disconnect in their distribution and were discovered to be sisters, and together they form a new higher order branch within the Nearctic oak clade (section Ponticae) interestingly both species share similarities in habit, as low growing clonal shrubs, and in general morphology. These species are likely relicts of an early branching of Quercus that has undergone a lot of range extinct. Estimates of divergence suggest a splitting time around 16-20 million years ago (Manos, 2016).
Another new lineage comes with resolving section Virentes as the sister to all white oaks (section Quercus) clarifying this group as a phylogenetically distinct branch of the Quercus tree. This section is one of the most distinctive groups, with a compelling geographic distribution that straddles from tropical zones to temperate ones (Hipp et al., 2018)
Within the Cerris-Ilex clade reconstructions based on cpDNA sequences separated Cerris from Ilex. However previous studies based on CRC sequences and on ITS suggested that the group Cerris and Ilex were mutually monophyletic. In fact based on fossil records of both groups and the results of dating by Hubert et al., 2014, Group Cerris seems to be a relatively recent off-shoot of group Ilex. This study is the first to show the subdivision of the new world clade into three well-supported subclades corresponding to the groups: Protobalanus, Lobatae and Quercus.
The sister group of the oaks appears to be the monotypic castaneoid genus Notholithocarpus (tanbark oak) of western North America that also bears an acorn-like fruit (Manos et al., 2008). In turn, this group is sister to a clade including the genera Castanea and Castanopsis. Notholithocarpus densiflorus is a paleoendemic species of the California Floristic Province, often occurring in the evergreen forests of the north Coast Ranges. This close relation of oaks to several castaneoid genera appears to confirm that the evolution of the acorn and the valveless cupule typically associated with Quercus is the result of the loss of lateral flowers in the dichasially arranged pistillate flowers. Interestingly, several independent losses of lateral flowers can be found in this clade as well as elsewhere in the family (Kremer et al., 2012).
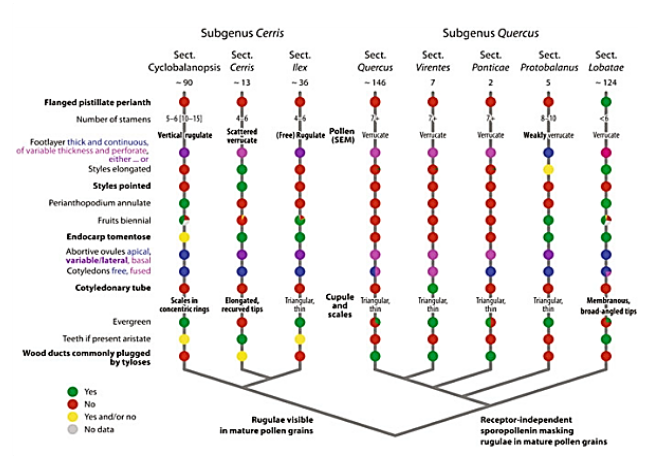
Revised sectional classification of oaks and diagnostic characters of lineages. The basic phylogenetic relationships of the six infrageneric groups of oaks are shown, formalized here as sections in two monophyletic subgenera, subgenus Cerris and subgenus Quercus. Section-specific traits in bold; subgenus-diagnostic traits indicated at the respective branches of the schematic phylogenetic tree. Note «yes» (green) and «no» (red) refers to whether the mentioned trait is observed or not in members of section but should not be generally viewed as derived or ancestral (Denk, et al., 2017).
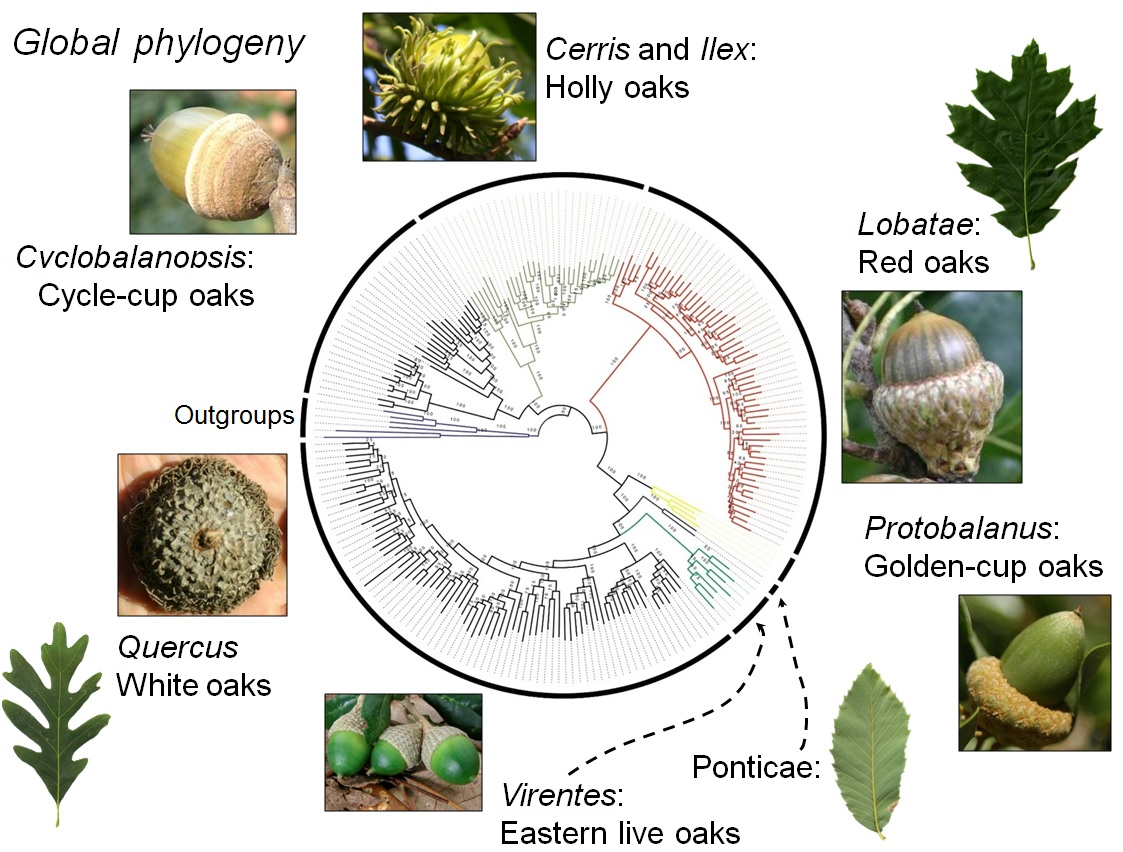
Global phylogeny. Image taken from Manos, 2017.